Gallery
Photos from events, contest for the best costume, videos from master classes.
 |  |
 |  |
 |  |
 |  |
 |  |
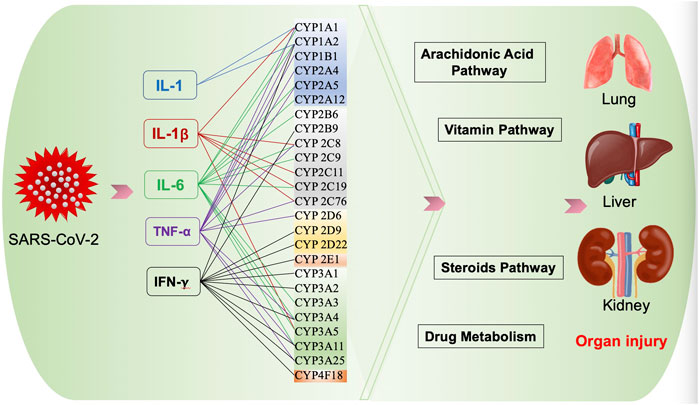 |  |
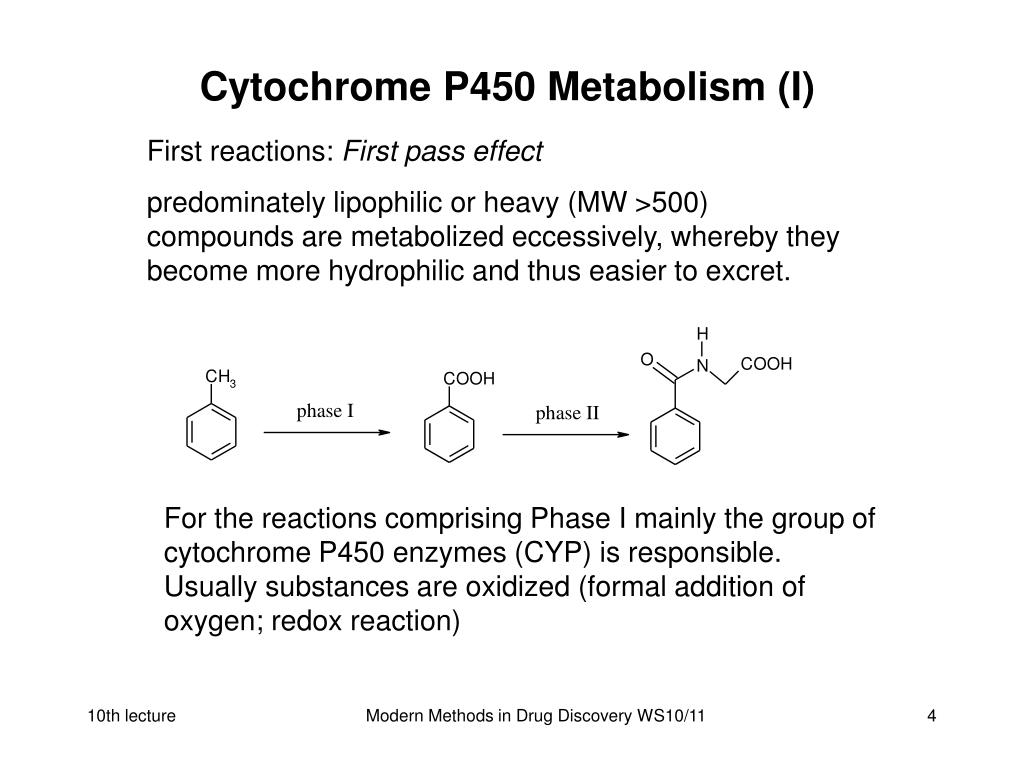
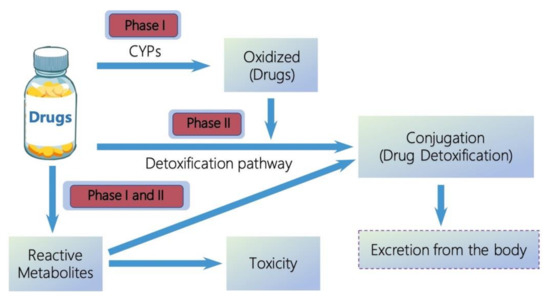
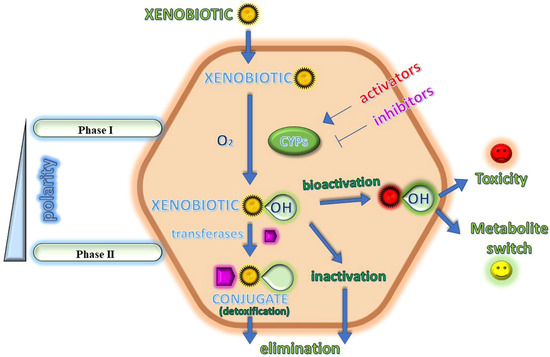
Metabolism. Gabapentin is not appreciably metabolized. Gabapentin enacarbil, the prodrug of gabapentin, is rapidly and efficiently converted to gabapentin by first-pass hydrolysis following oral administration. Elimination Route. Excreted renally as unchanged drug. Half-life. Approximately 5–7 hours. Special Populations Metabolism and excretion They are not metabolised by the liver and do not affect the cytochrome P450 system, major cytochrome P450 system isoenzymes; however, drug-induced hepatotox-icity has been described in case reports.16 Elimination is mostly done by the kidney and is proportional to the Figure 1. Structure of GABA: gabapentin and Gabapentin is eliminated from the systemic circulation by renal excretion as unchanged drug. Gabapentin is not appreciably metabolized in humans. Gabapentin elimination half-life is 5 to 7 hours and is unaltered by dose or following multiple dosing. Gabapentin elimination rate constant, plasma clearance, and renal clearance are directly Gabapentin is not bound to plasma proteins, does not induce hepatic enzymes and is not metabolized. At steady state, it has a half-life of 6-8 h, and is eliminated unchanged by renal route with a plasma clearance proportional to the creatinine clearance. Purpose Tamoxifen undergoes metabolic activation by cytochrome P450 (CYP) enzymes to metabolites with more potent anti-estrogenic effects. Numerous studies demonstrate decreased tamoxifen efficacy associated with reduced CYP2D6 activity or lower Z-endoxifen concentrations. Women taking tamoxifen frequently experience vasomotor symptoms (VMS) that may require medical treatment. Many medications Metabolism: In humans, gabapentin undergoes minimal metabolic alteration, largely retaining the original structure. Gabapentin does not induce or inhibit CYP enzymes. Also, none of the CYP enzyme inhibitors alter their pharmacokinetics. As a drug, gabapentin was formerly considered as a structural analogue of the inhibitory neurotransmitter γ-aminobutyric acid (GABA). However, preliminary studies proposed that gabapentin did not bind to either GABA-A or GABA-B receptors 11, nor was it transform metabolically into GABA. 12 Phase I Metabolism: Phase II Metabolism: CYP 1A2: CYP 2B6: CYP 2C8: CYP 2C9: CYP 2C19: CYP 2D6: CYP 3A4: CYP 2E1: UGT 2B7 & UGT 1A9: Ketorolac: X: X: X: Salsalate Meloxicam Gabapentin & Pregabalin Duloxetine Carbamazepine & Oxcarbazepine Desipramine Topiramate Weak Inhibitor Weak Inducer Acetaminophen Baclofen Tizanidine: X Fentanyl They are not metabolised by the liver and do not affect the cytochrome P450 system, major cytochrome P450 system isoenzymes; however, drug-induced hepatotoxicity has been described in case reports. 16 Elimination is mostly done by the kidney and is proportional to the creatinine clearance. Accumulation can cause renal failure resulting in This is a list of cytochrome P450 modulators, Gabapentin; Isoniazid; Ketoconazole; "MEDICATIONS METABOLIZED BY CYTOCHROME P450 3A4" The aim of this paper is to review a number of new antiepileptic agents (i.e. felbamate, gabapentin, lamotrigine, levetiracetam, oxcarbazepine, tiagabine, topiramate, vigabatrin and zonisamide) for their inducing and/or inhibitory properties in humans, mainly considering the interactions where they gabapentin enacarbil (GEn) to the α2δ subunit corresponds to the treatment of restless leg syndrome (RLS) symptoms. Indication: GEn is FDA approved for treatment of RLS. Pharmacokinetics: GEn is primarily excreted by the kidneys and neither gabapentin nor GEn is substrate, inhibitor, or inducer of the major CYP450 system. Due to a lack of hepatic metabolism, and a low degree of protein binding, gabapentin has very limited potential for drug interactions. In addition, gabapentin does not inhibit or induce the major cytochrome P450 enzymes responsible for drug metabolism. Ethosuximide, gabapentin, tiagabine, and vigabatrin are neither inducers nor inhibitors of drug metabolism. Hepatic enzyme inhibition usually occurs because of competition at the enzyme site. Knowledge of the specific metabolic enzymes involved in the metabolism of AEDs allows clinicians to predict potential interactions. High volume of distribution (58L), less than 3% of gabapentin is bound to plasma proteins : Metabolism: Not metabolized to a significant extent in humans. Elimination: Solely renal excretion as unchanged drug, and can be removed from plasma by hemodialysis. Half-life As a result, metabolism-related factors do not necessitate dosage alterations of gabapentin and concomitant AEDs after prolonged therapy. The drug is excreted unchanged in urine; plasma clearance is linearly related to creatinine clearance; and dosage is readily adjusted based on renal function. Unlike many of the older drugs, lamotrigine, vigabatrin and gabapentin have a predominantly renal excretion and are not metabolised through the cytochrome P450 system. They do not induce their own metabolism or that of other commonly used anticonvulsants. Because metabolism occurs via successive CYP450 enzymes, zolpidem has a high potential for drug–drug interactions with agents that inhibit or induce CYP450 enzymes. Although gabapentin is not metabolized via the CYP450 pathway , there is potential for co-administration with gabapentin, and therefore, any potential effects of zolpidem on the In general, newer AEDs have more predictable kinetics and lower risks for drug interactions. This is because many are minimally or not bound to serum proteins, are primarily renally cleared or metabolized by non–cytochrome P450 isoenzymes, and/or have lower potential to induce/inhibit various hepatic enzyme systems. Gabapentin is not appreciably metabolized in humans. Gabapentin elimination half-life is 5 to 7 hours and is unaltered by dose or following multiple dosing. Gabapentin elimination rate constant, plasma clearance, and renal clearance are directly proportional to creatinine clearance.
Articles and news, personal stories, interviews with experts.
Photos from events, contest for the best costume, videos from master classes.
 |  |
 |  |
 |  |
 |  |
 |  |
 |  |